Cell Proliferation and Cell Viability Assay Services
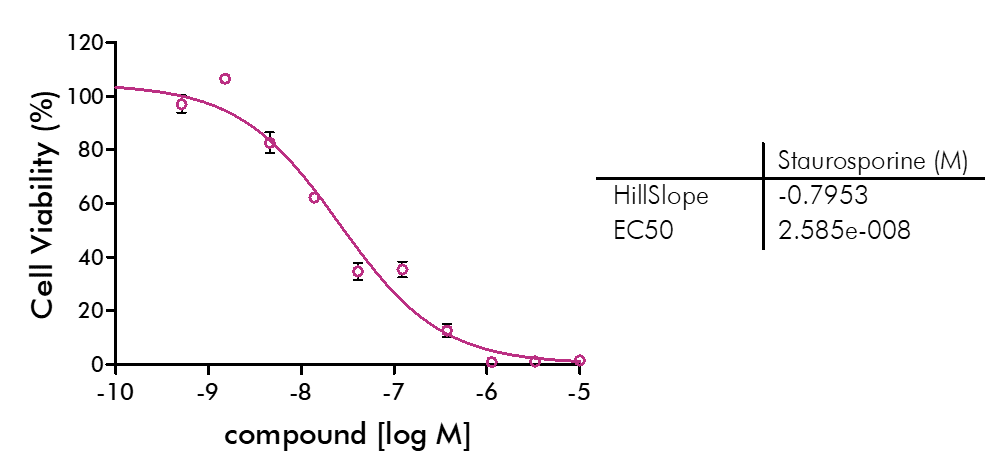
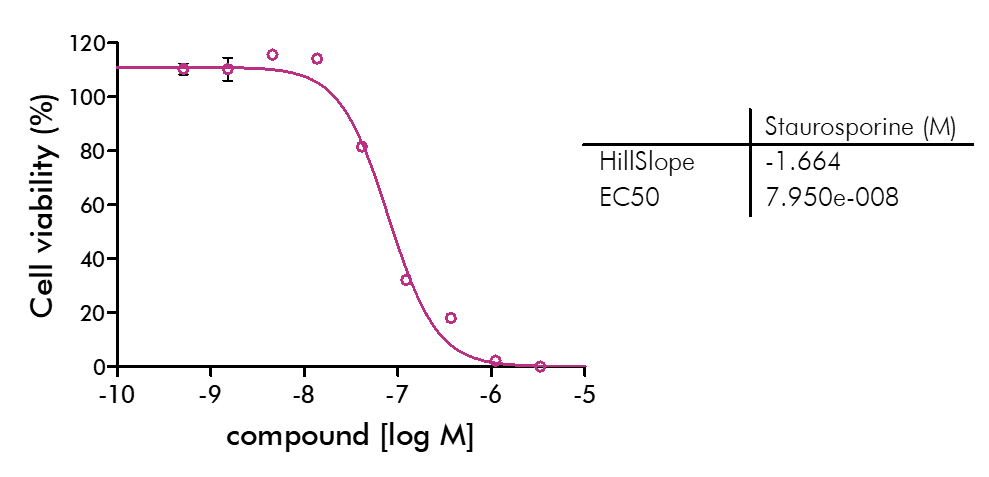
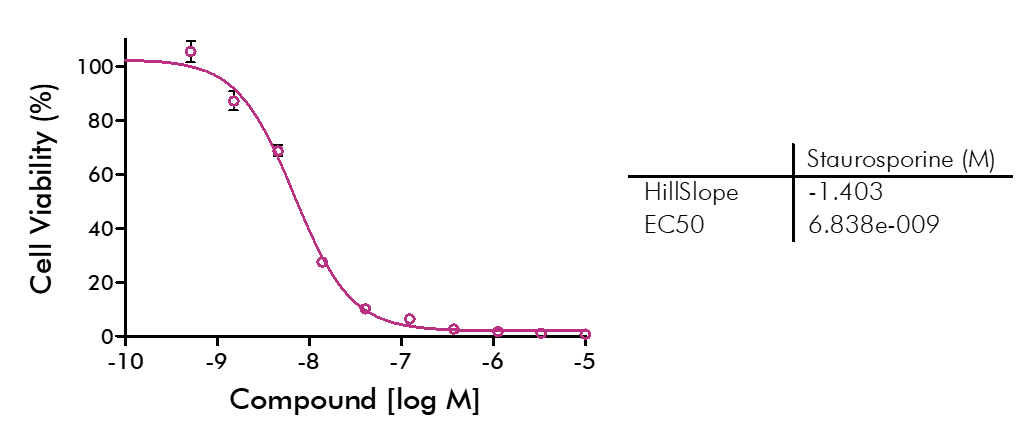
Cell viability and cell proliferation assays are indispensable for any cancer drug discovery project. Cell proliferation assays are widely used as an initial test for the therapeutic efficacy of anti-cancer agents.
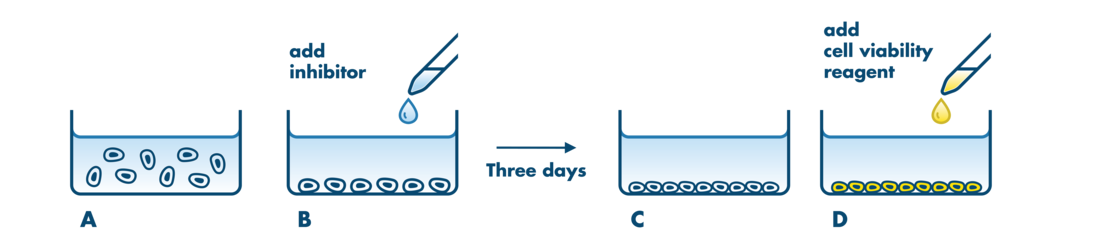
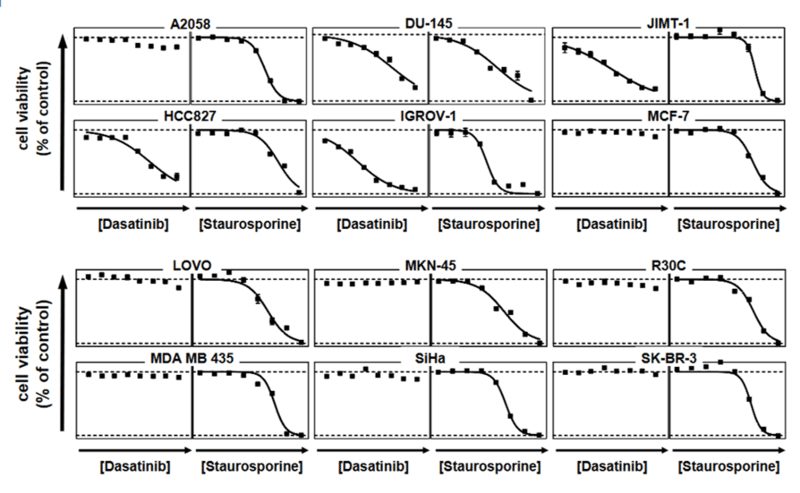
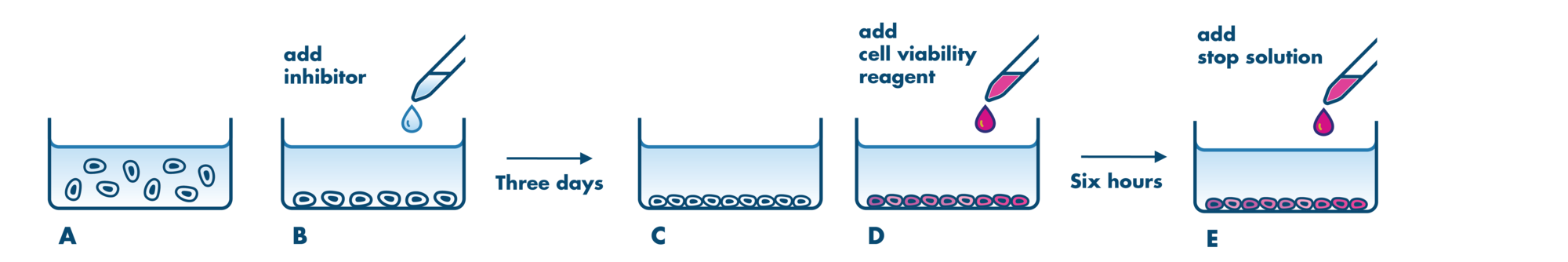
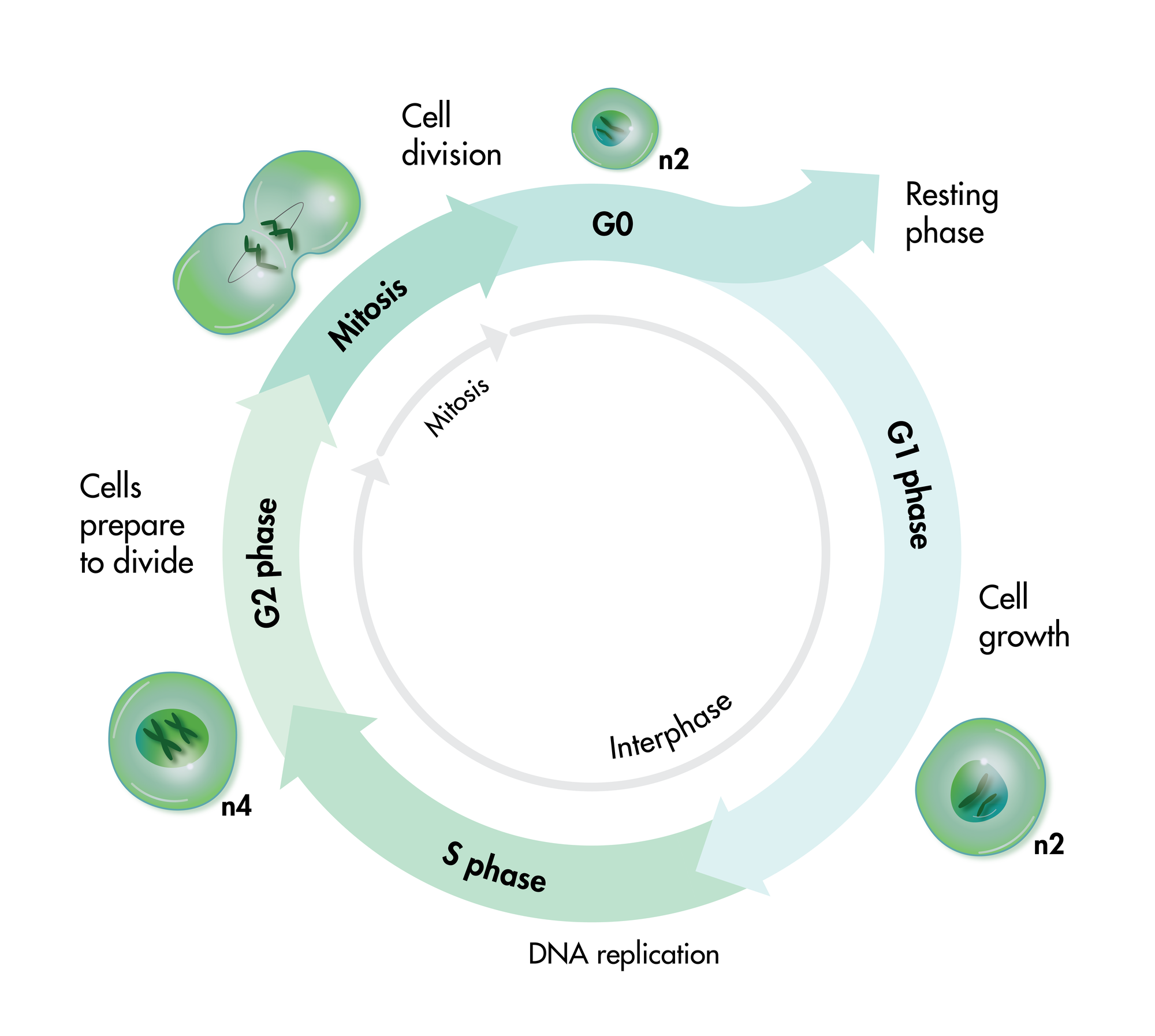
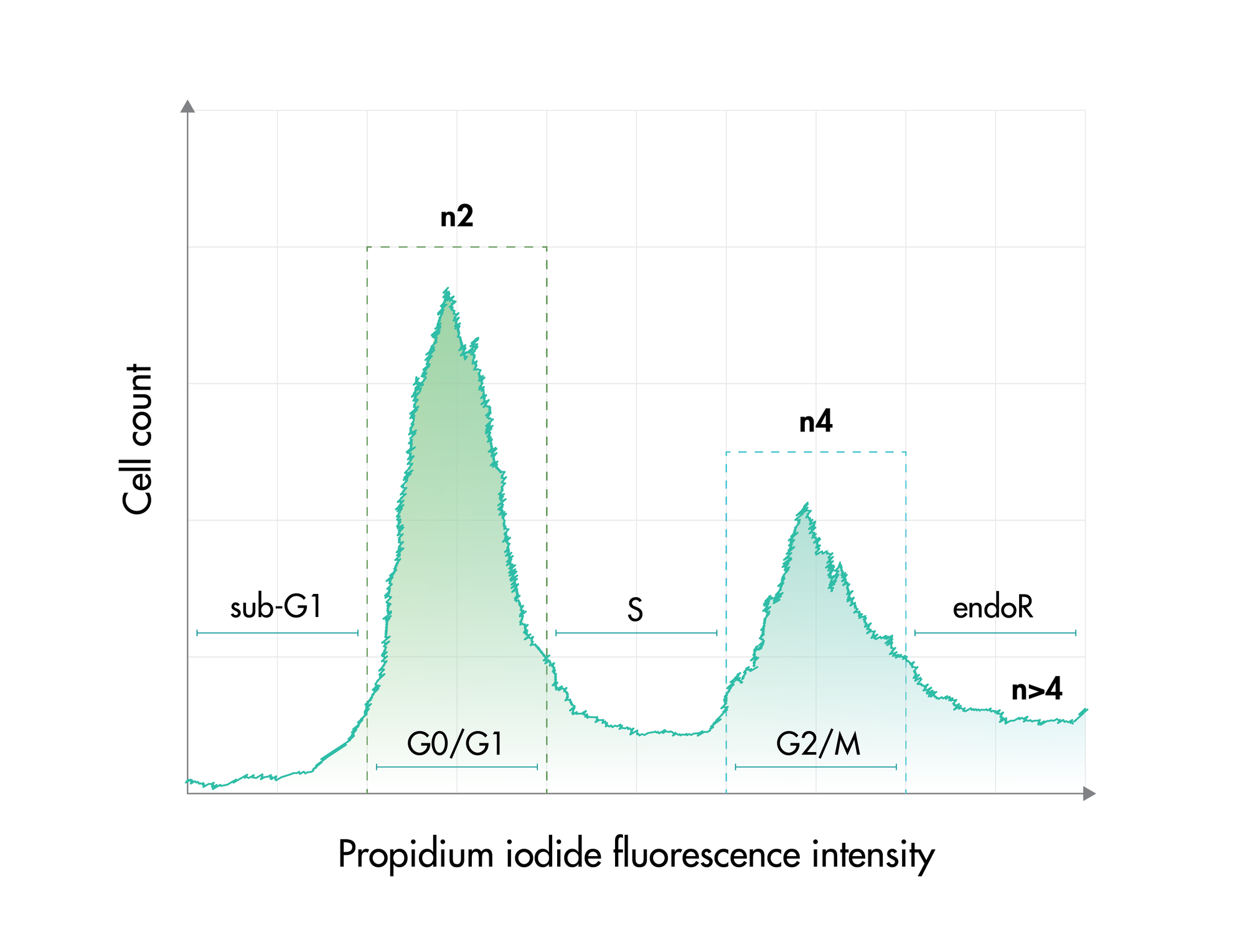
For analysis of the impact of anti-cancer drugs, the most common cellular phenotypic assay is the 2D cell proliferation assay in which tumor cells are growing in suspension culture or as monolayers in cell culture plates. Our standard assay format Cell Titer-Glo which allows the testing of large numbers of compounds in a fast and cost-efficient fashion. Due to its simplistic setup, the cell proliferation assay is generally regarded as a pretest to more sophisticated assays such as apoptosis or 3D soft agar assays, which are also available at Reaction Biology.
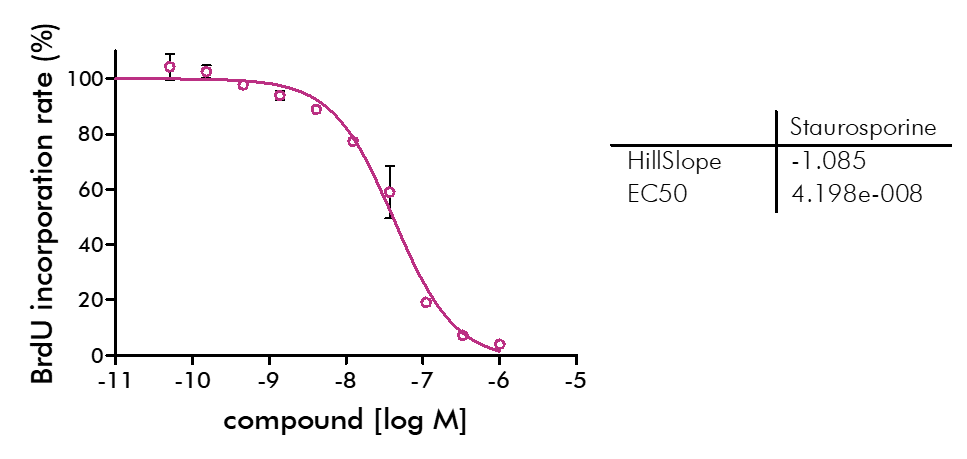
- Beside Cell Titer-Glo, we offer potency testing of new drugs on cell proliferation and viability with a variety of alternative assay formats are available including ATPlite, CyQUANT, BrdU incorporation, MTT, Alamar Blue, and WST-8 readouts as well as real-time kinetics measured via IncuCyte.
- Regular large tumor cell panel screening is available with our ProLiFiler™ screening service.
- Our 2D cell proliferation assays allow for drug incubation times up to 3 days. For projects that may require incubation times >3 days to develop a drug response, we recommend our 3D Tumor Spheroid Assay Service or the Soft Agar Assay Service which are well suited to this application. Please contact us to discuss the details.
Our gold standard assay formats, the large cell line panel, and exceptional service make us the choice contract research organization for quality cell proliferation assays. Contact our global business development team today to discuss your contract research needs.